Alessandro Gardini, Ph.D.
-
Associate Professor, Genome Regulation and Cell Signaling Program, Ellen and Ronald Caplan Cancer Center
Gardini studies the epigenetic control of transcription during cell differentiation and oncogenesis.
Born and raised in Italy, Gardini obtained a B.S./M.S. in medical biotechnology at the University of Bologna and attended the graduate school of Molecular Medicine at the University of Milan. He trained as a postdoctoral fellow with Dr. Ramin Shiekhattar at the Center of Genomic Regulation in Barcelona, The Wistar Institute and the University of Miami Medical School. He joined Wistar as an assistant professor in 2015. Gardini is a scholar of the Leukemia Research Foundation and the American Cancer Society.
The Gardini Laboratory

The Gardini Laboratory
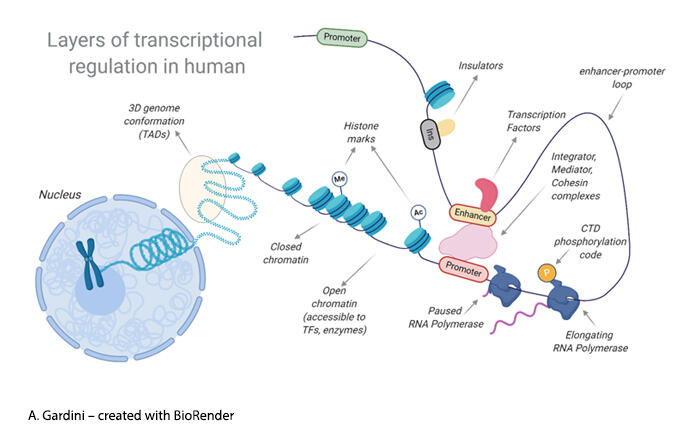
Transcriptional regulation is a multi-layer process carried out by specialized transcription factors, co-factors and enzymatic machineries working within the context of a structured chromatin landscape.
We use a combination of genomics, proteomics and biochemistry to investigate how basal transcriptional complexes and epigenetic modulators orchestrate gene expression in normal and tumor cells.
-
Associate Staff Scientist
Sandra Deliard, Ph.D.
-
Graduate Students
Kelsey Leach
Joel Lee
Sarah Offley (UPenn-CAMB)
-
Alumni
Marco Trizzino, Ph.D. (2016-2019), assistant professor, Thomas Jefferson University
Elisa Barbieri, Ph.D. (2015-2019), Marie Sklodowska-Curie Fellow, University of Edinburgh
-
Learn about job opportunities at The Wistar Institute here.
Research

We dissect the transcriptional mechanisms that control fate choice and differentiation of human hematopoietic cells, particularly in the myeloid compartment. We are especially interested in the role of transcriptional enhancers.
Enhancers are distal regulatory elements, scattered throughout the entire genome, that direct gene regulation during development. Furthermore, enhancers are active spots for transcription of long noncoding eRNAs (Gardini and Shiekhattar, 2015). These short-lived, non-polyadenylated transcripts are required to establish a link between the promoter and the distant regulatory enhancer (chromosomal looping) and contribute to the expression of the target protein-coding gene. The underlying mechanisms are poorly understood and the potential contribution of enhancers and eRNAs to cancer has just started to emerge. The lab is keen on understanding how enhancer networks get activated and promote differentiation of myeloid cells, and how this process is disrupted during leukemogenesis. We recently uncovered a novel enhancer regulatory axis in myeloid progenitor cells (Barbieri, Trizzino et al., 2018), centered around the EGR-1 transcription factor and a newly characterized module of the Integrator complex.
Additional studies (Trizzino, Zucco et al., 2021) revealed that EGR-1 also coordinates a repressive activity, independent of Integrator, in mature macrophages. Particularly, EGR-1 restrains the activity of inflammatory enhancers and effectively curbs the response of macrophages to inflammatory stimuli.

Eukaryotic transcription pivots around the balanced activity of kinases and phosphatases that regulate the transcription initiation, early elongation and termination checkpoints. While the dynamic recruitment of kinases (i.e. CDK7, CDK9) and their cognate cyclins have been studied for many years, the association of phosphatases with active transcription has been largely understudied. We recently uncovered that protein phosphatase 2A (PP2A) is recruited at the pause-release checkpoint where it opposes CDK9 activity (Vervoort, Welsh et al., 2021). We found that the Integrator subunit INTS6 draws PP2A to the transcription bubble to dephosphorylate the RNAPII-CTD (especially Ser2) and maintain polymerase in a paused conformation. PP2A activity is frequently lost in cancer and restoring its ability to curb transcription opens up new therapeutic strategies for tumors ‘addicted’ to transcriptional elongation.

Integrator is a large, evolutionarily conserved, multiprotein complex that is quickly emerging as a centerpiece of transcriptional regulation in higher eukaryotes (Welsh and Gardini, 2022). The Integrator complex is a regulatory hub for several transcriptional and epigenetic processes. For instance, Integrator’s INTS11 subunit controls release of RNA Polymerase II at promoters via endonucleolytic cut of nascent RNA (Gardini et al., 2014; Beckedorff et al., 2020). In addition, INTS11 regulates enhancer function through the processing of noncoding eRNAs (Lai, Gardini et al., 2015). We identified a new functional module of Integrator, centered around the INTS13 subunit, that prompts enhancer activation in myeloid cells (Barbieri, Trizzino et al., 2018). More recently, we showed that the INTS6/INTS8 subunits of the complex recruit the PP2A phosphatase to chromatin, to control phosphorylation of the RNAPII-CTD as well as other polymerase co-factors implicated in pausing and elongation (Vervoort, Welsh et al., 2021). Notably, Integrator subunits are found mutated in developmental diseases and are frequently lost in certain tumor types. The lab keeps pursuing the biochemical and functional dissection of all 14 subunits of the Integrator complex.

Conventional transcriptomic analyses (i.e. ribo-depleted RNA-seq, 3’-seq) offer a glimpse into global gene regulation but often fail to convey the most accurate picture. Because RNA-seq captures steady-state and cytoplasmic transcripts, changes in gene expression can be hindered by RNA stability. Furthermore, RNA Polymerase dynamics is only revealed by real-time nascent RNA transcription. We strive to implement and optimize techniques of nascent RNA-seq, improving their reproducibility and applicability. To this purpose, we recently developed fastGRO (Barbieri, Hill et al., 2020) as a global nuclear run-on assay of rapid execution and suitable for primary cells.
Chromatin remodelers regulate DNA accessibility by positioning nucleosomes at active promoters and enhancers. We investigate the function of SWI/SNF, the most mutated chromatin remodeler across all human cancers. Subunits of SWI/SNF, such as ARID1A, are found mutated in sporadic ovarian tumors and their specific contribution to SWI/SNF activity is poorly understood. We recently elucidated a novel function of ARID1A in pausing of RNA Polymerase II (Trizzino et al., 2018).
Gardini Lab in the News
Selected Publications
The PP2A-Integrator-CDK9 Axis Fine-tunes Transcription and Can Be Targeted Therapeutically in Cancer.
Vervoort, S.J., Welsh, S.A., Devlin, J.R., Barbieri, E., Knight, D.A., Offley, S., Bjelosevic, S., Costacurta, M., Todorovski, I., Kearney, C.J., et al. “The PP2A-Integrator-CDK9 Axis Fine-tunes Transcription and Can Be Targeted Therapeutically in Cancer.” Cell. 2021 May 17; doi: 10.1016/j.cell.2021.04.022
EGR1 is a Gatekeeper of Inflammatory Enhancers in Human Macrophages
Trizzino, M., Zucco, A., Deliard, S., Wang, F., Barbieri. E., Veglia, F., Gabrilovich, D., Gardini, A. “EGR1 is a Gatekeeper of Inflammatory Enhancers in Human Macrophages” Sci Adv. 2021 Jan 13;7(3):eaaz8836. doi: 10.1126/sciadv.aaz8836. Print 2021 Jan.
Rapid and Scalable Profiling of Nascent RNA with fastGRO
Barbieri, E., Hill, C., Quesnel-Vallières, M., Zucco, A.J., Barash, Y., Gardini, A. “Rapid and Scalable Profiling of Nascent RNA with fastGRO” Cell Rep. 2020 Nov 10;33(6):108373. doi:10.1016/j.celrep.2020.108373.
Targeted enhancer activation by a subunit of the Integrator complex
Barbieri, E., Trizzino, M., Welsh, S.A., Owens, T.A., Calabretta, B., Carroll, M., Sarma, K., Gardini, A. “Targeted enhancer activation by a subunit of the Integrator complex.” Mol Cell. 2018.
The tumor suppressor ARID1A controls global transcription via pausing of RNA Polymerase II.
Trizzino, M., Barbieri, E., Petracovici, A., Wu, S., Welsh, S.A., Owens, T.A., Licciulli, S., Zhang, R., Gardini, A. “The tumor suppressor ARID1A controls global transcription via pausing of RNA Polymerase II.” Cell Reports. 2018.